The growing interest in genomic research, personalized medicine and ribonucleic acid (RNA)-based therapies is fueling demand for high quality oligonucleotides.
RNA is a biological molecule that plays a crucial role in the transfer of genetic information from deoxyribonucleic acid (DNA) to ribosomes during protein synthesis. RNA therapeutics is a rapidly growing class of medications that will revolutionize the way many diseases are treated and make personalized medicine a reality.
Oligonucleotides, meanwhile, are short, synthesized pieces of DNA that are becoming increasingly important in research, diagnostics, and gene editing, with some of the better-known applications being in the polymerase chain reaction (PCR) tests used to detect COVID-19, and as therapeutic agents that target specific genes to alter their behavior, making them of particular interest to treat cancer, genetic conditions and rare diseases.
Even though the first oligonucleotides were chemically synthesized in the late 1970s, it is recent advances in the field, such as clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9), and improvements in the ability to synthesize longer and more complex oligonucleotides that have led to a surge in this rapidly expanding field. As a result, there is a growing demand for advanced tools and methods that can accurately verify oligonucleotide sequences as well as identify related impurities.
The importance of accurate sequencing in RNA and oligonucleotide analysis
The sequence accuracy of RNA and oligonucleotides is important as even a single nucleotide change can significantly alter their function and specificity.[i] RNA silencing is a mechanism by which cells regulate gene expression by degrading or inhibiting the translation of messenger RNA (mRNA) molecules. This important regulatory mechanism is used in many cellular processes, including development, differentiation, and response to viral infection.
As a result, a correct primary sequence is crucial for proper folding and stability, which are necessary for its biological function. Any sequence inaccuracies can result in off-target effects which can have serious consequences for the cell or organism.
Researchers at Axolabs, a leading custom research organization (CRO) specializing in the development of DNA- and RNA-based therapeutics, have emphasized that accuracy is essential to ensure the molecular identity and to avoid contaminations with potential risk for efficacy and potency. Avoiding impurities and product degradation during production is critical to ensuring that the biopharmaceutical products they produce meet regulatory and approval requirements.
Typical sequencing methods
Oligonucleotides can be sequenced using various methods. The most widely used method is Sanger sequencing, developed by Frederick Sanger in the 1970s. This involves the sequential addition of nucleotides to a growing DNA strand, and the addition of modified nucleotides called dideoxy nucleotides. These additions stop the extension of the DNA chain, generating a series of DNA fragments of varying lengths that are then separated by size to enable reading. Once the DNA sequence is obtained, it needs to be annotated to identify genes and other functional elements such as the location and structure of genes, as well as regulatory regions such as promoters and enhancers. Next, RNA sequencing is used to measure the expression of genes in a sample. The resulting sequence reads are assembled to create a transcriptome, and differential gene expression analysis is performed to identify genes that are differentially expressed between different samples or conditions.
The technique that has been making the most headlines in recent years is next-generation sequencing (NGS), as it is capable of simultaneously sequencing millions of DNA fragments in parallel, generating vast amounts of data that can be used to identify genetic variations. However, it does not offer a comprehensive characterization and is most useful for simple oligos and RNA molecules.
Single molecule real-time sequencing on the other hand, sequences individual DNA molecules, allowing for longer reads and improved accuracy. This presents another challenge – because oligonucleotides are so complex, it is only possible to accurately analyze a couple of samples a day.
Liquid chromatography (LC) is often used in combination with mass spectrometry (MS) to sequence oligonucleotides. It is used to separate and identify individual oligonucleotides based on their physical and chemical properties, such as size and charge. Though, this technique does not always yield enough information, especially on impurities and by-products.
Oligonucleotide sequencing with tandem mass spectrometry (MS/MS)
MS has great appeal in the field of oligonucleotide-based therapeutics because it directly measures the mass-to-charge ratio of the molecule to directly obtain the molecular weight of the target. MS provides the robustness, high selectivity and sensitivity required for testing virtually any system, analyzing single- and double-stranded oligonucleotides. In addition, it enables the quantitation of synthesis by-products.
Quadrupole time of flight mass spectrometry (QTOF-MS) instruments are being used to characterize highly modified oligonucleotide sequences as they can determine monoisotopic masses of intact oligonucleotides and their fragment ions with high mass accuracy. The high isotopic fidelity of these instruments, combined with the established SNAP algorithm for monoisotopic peak picking, provide reliable annotation of precursor and fragment ion masses (OligoQuest, Bruker). MS and MS/MS spectra are thereby matched with high certainty to sequence candidates such as the target sequence and expected by-products.
MS/MS is a two-step technique used to identify a sample. In the first step a precursor ion of the molecule of interest is isolated. In a second step, this isolated ion is fragmented into smaller ions and these fragment ions are measured. This fragmentation step provides detailed information on the structure of the sample, providing further structural information.
This method is used to determine the sequence, length, and modification status of oligonucleotides. This is valuable information for quality control, process development, and stability analysis of drugs and diagnostics based on oligonucleotides. As a result, MS/MS has become a widely adopted tool for the analysis of oligonucleotides and RNA due to its capability to identify and quantify fragment ions with high sensitivity. Currently, though, data interpretation requires a high level of experience and is very time-consuming, creating a gap between fast data acquisition and slow manual interpretation.
New software advances are bridging the MS/MS data gap by adding powerful tools for the interpretation of RNA and oligonucleotide data. Proven algorithms for peak detection and deconvolution of MS and MS/MS data obtained with high isotopic fidelity ensure correct mass assignments even in samples covering a high dynamic range (Figure 1).
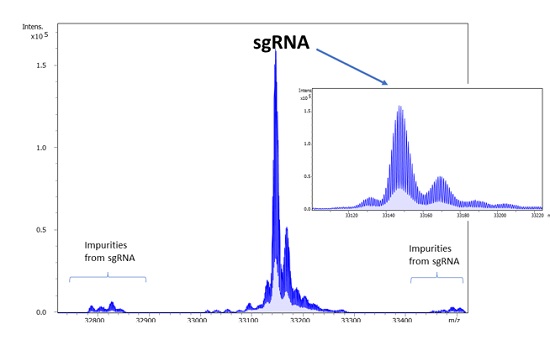
Figure 1: Deconvoluted spectrum for a sgRNA and associated low level impurities.
Using software to simplify analysis
The complexity of the data captured for sequence verification means that manual interpretation is expensive and time-consuming, and requires in-depth, technical know-how. To facilitate the analysis of such large datasets, tools have been developed that use algorithmic approaches to analyze data quickly and reliably.
For example, an annotation engine can be used to automatically identify oligonucleotide fragments, allowing rapid analysis of complex MS/MS data, and reporting the results back to the researcher in a readable format, see example in Figure 2.
To make it easier to interpret results, software tools are used to process raw data into a usable format and provide easy-to-read, graphical representations of the data. They can also manage and store large amounts of data, to simplify the comparison of results over time.
Repetitive tasks can be automated to reduce the risk of human error, and
pre-programmed, proven algorithms can ensure correct mass assignments and the validation of oligonucleotides using relevant datasets. The software can be programmed to perform specific, targeted analyses, thereby increasing the specificity of results and reducing the risk of false positive results. The Axolabs team describes how that capability streamlines their workflows.
The SNAP algorithm in OligoQuest helps Axolabs with selecting the mono-isotopic signal of one fragment, reducing the probability of mistakes. Additionally, OligoQuest’s improved usability makes it much more efficient as it provides an overview of complex data with one graphic, this data visualization brings the results to life.
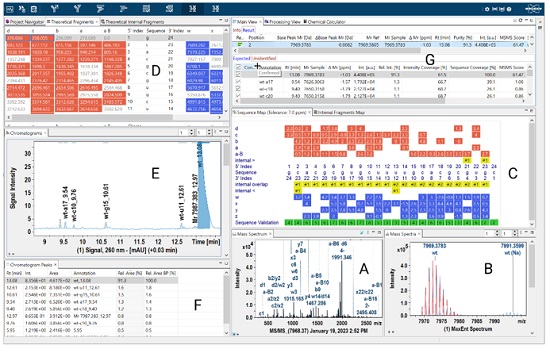
Figure 2: Example output of MS/MS spectrum of an 2´-O-permethylated RNA 24-mer with fragment ion annotations in Bruker’s BioPharma Compass software (A) and the intact mass annotated and overlayed with the theoretical isotope pattern (B). The Sequence Map (C) displays the 5´- (red) and 3´-fragments (blue) including the observed ppm mass errors. The theoretical fragment ion masses including the matches with the spectrum are also displayed (D). The chromatogram (E) and the chromatographic peak table with the quantitative UV peak data are shown in (F). The analysis is summarized in tabular form (G).
The future of oligonucleotide characterization and RNA therapeutics
Oligonucleotide and RNA therapeutics are thought to have nearly unlimited capacity to address unmet clinical needs,[ii] and are destined to change the standard of care for many diseases including the fight against cancer.
The number of RNA drugs under development and in clinical trials is growing rapidly, and this is largely due to advances in MS and other sequencing techniques that are enabling more accurate and sensitive analysis. This will further increase their demand in biotech and molecular biology, particularly as growth in the field of genomics and research into personalized therapies continues. Meanwhile, new RNA-targeted drugs are being developed to target specific RNA molecules and disrupt disease-causing processes at the RNA level.
The development of new methods for synthesizing and delivering oligonucleotides and RNA therapeutics that are cheaper and more efficient, along with further advances in gene editing technology such as CRISPR-Cas9, will continue to drive demand for these therapeutics and their associated tools.
Authors
Timo Schierling, Scientist (Analytics R&D), Axolabs GmbH
Julia Schneider, Principal Research Associate, Axolabs GmbH
Stephan Seiffert, Team Leader (Method Development Analytics), Principal Scientist, Axolabs
Christian Albers, Business Development Manager Pharmaceuticals Market, Bruker Daltonics
Reference
[i] Bruce Alberts, et al, From RNA to Protein, Molecular Biology of the Cell 4th edition, 2002
[ii] Tulsi Ram Damase et al, The Limitless Future of RNA Therapeutics, Front. Bioeng. Biotechnol.,Vol 9, 2021