The convergence of genomics, proteomics, transcriptomics, metabolomics, and pathology data represents the most significant advancement in precision medicine since the completion of the Human Genome Project. This multi-omic data integration precision medicine approach promises to revolutionize healthcare by providing unprecedented insights into disease mechanisms, treatment responses, and patient outcomes. As we advance toward an era of truly personalized medicine, the ability to synthesize diverse biological data layers is becoming essential for delivering optimal patient care and developing next-generation therapeutic strategies.
The Evolution of Multi-Omic Integration in Precision Medicine
Multi-omic data integration precision medicine has evolved from a theoretical concept to a practical reality through advances in high-throughput sequencing, mass spectrometry, and computational biology. The traditional approach of analyzing single data types in isolation has given way to sophisticated integration methodologies that capture the complex interplay between genetic variation, gene expression, protein abundance, metabolic profiles, and tissue architecture. This holistic view enables researchers and clinicians to understand disease processes at multiple biological levels simultaneously.
The shift toward integrated approaches reflects growing recognition that single-omic analyses often fail to capture the full complexity of human disease. Genomic data alone, while informative about predisposition and risk factors, cannot fully predict how diseases will manifest or progress in individual patients. Similarly, proteomic or metabolomic data in isolation may miss critical regulatory mechanisms that operate at the genetic or transcriptional level. Multi-omic data integration precision medicine addresses these limitations by combining complementary data types to create comprehensive molecular portraits of health and disease.
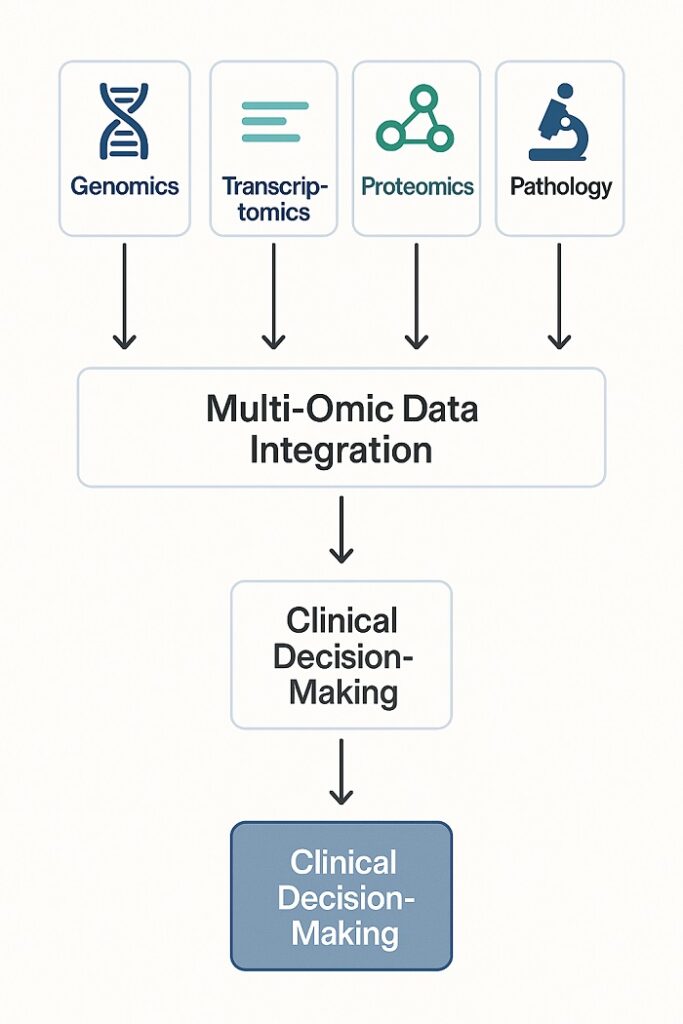
Multi-Omic Integration Workflow showing data flow to clinical decision-making
Recent technological advances have made multi-omic integration increasingly feasible and cost-effective. Next-generation sequencing costs have decreased dramatically while throughput has increased exponentially. Mass spectrometry-based proteomics and metabolomics have achieved greater sensitivity and reproducibility. Digital pathology platforms enable quantitative analysis of tissue architecture and cellular organization. These improvements have created opportunities for large-scale multi-omic studies that would have been impossible just a decade ago.
Genomics as the Foundation Layer
Genomic data serves as the foundational layer for multi-omic data integration precision medicine approaches, providing the blueprint from which all other molecular processes emerge. Whole genome sequencing reveals structural variants, copy number alterations, and single nucleotide polymorphisms that influence disease susceptibility and treatment response. Targeted gene panels enable focused analysis of clinically relevant mutations while maintaining cost-effectiveness for routine clinical applications.
The integration of germline and somatic genomic data creates comprehensive genetic profiles that inform both inherited disease risk and acquired pathogenic processes. Pharmacogenomic analysis reveals how genetic variants affect drug metabolism, efficacy, and toxicity, enabling personalized medication selection and dosing. Tumor genomic profiling identifies driver mutations and therapeutic targets that guide precision oncology treatment decisions.
Advanced genomic analysis techniques, including structural variant detection, loss of heterozygosity analysis, and mutational signature analysis, provide deeper insights into disease mechanisms and evolution. These approaches are particularly valuable in oncology, where tumor heterogeneity and clonal evolution complicate treatment selection. Multi-omic data integration precision medicine leverages genomic data as the stable reference point against which dynamic molecular changes can be interpreted.
Transcriptomics: Bridging Genotype and Phenotype
Transcriptomic analysis occupies a crucial position in multi-omic data integration precision medicine by revealing how genetic information is translated into functional molecular processes. RNA sequencing captures global gene expression patterns, alternative splicing events, and novel transcripts that may not be apparent from genomic analysis alone. Single-cell RNA sequencing provides unprecedented resolution of cellular heterogeneity and developmental trajectories within tissues and tumors.
The dynamic nature of transcriptomic data enables monitoring of disease progression and treatment response over time. Changes in gene expression patterns can indicate therapeutic efficacy, resistance development, or disease recurrence before these become apparent through conventional clinical assessments. Pathway analysis of transcriptomic data reveals dysregulated biological processes and potential therapeutic targets that may not be evident from individual gene expression changes.
Integration of transcriptomic data with genomic information enables identification of expression quantitative trait loci, revealing how genetic variants influence gene expression levels. This information is crucial for understanding the functional consequences of genetic variants and predicting their clinical significance. Multi-omic data integration precision medicine uses transcriptomic data to bridge the gap between static genomic information and dynamic phenotypic manifestations.
Proteomics: Functional Molecular Readouts
Proteomic analysis provides direct measurement of the functional molecules that execute biological processes, making it an essential component of multi-omic data integration precision medicine strategies. Mass spectrometry-based proteomics can quantify thousands of proteins simultaneously, revealing post-translational modifications, protein interactions, and pathway activities that are not apparent from gene expression data alone.
The integration of proteomics with genomics and transcriptomics enables identification of biomarkers with enhanced specificity and clinical utility. Protein abundance often correlates poorly with mRNA expression due to post-transcriptional regulation, protein stability, and degradation processes. Multi-omic approaches that combine these data types provide more accurate predictions of protein function and therapeutic response than either approach alone.
Phosphoproteomics analysis reveals signaling pathway activities and kinase networks that are critical for understanding disease mechanisms and identifying therapeutic targets. These dynamic modifications reflect cellular responses to environmental stimuli and therapeutic interventions, providing real-time insights into biological processes. Multi-omic data integration precision medicine uses phosphoproteomic data to identify actionable targets and monitor treatment responses at the molecular level.
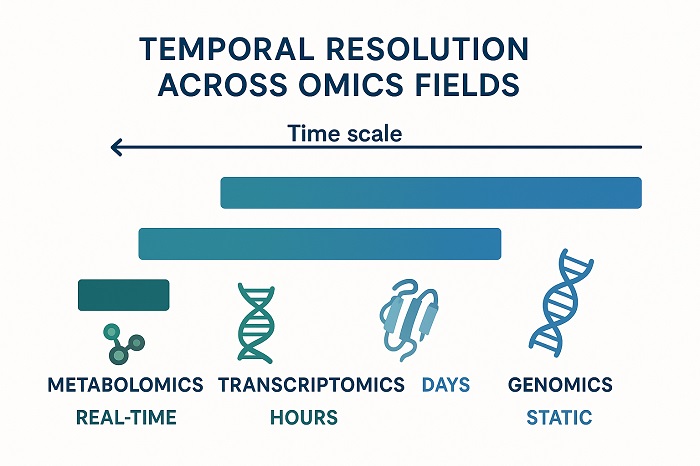 Temporal Resolution Comparison showing time scales of different omic measurements
Temporal Resolution Comparison showing time scales of different omic measurements
Metabolomics: Real-Time Cellular Physiology
Metabolomics provides the closest functional readout of cellular physiology, capturing the end products of genetic, transcriptomic, and proteomic processes. Small molecule metabolites reflect the integrated output of multiple biological pathways and provide sensitive indicators of disease states and treatment effects. Multi-omic data integration precision medicine incorporates metabolomic data to understand how upstream molecular changes translate into altered cellular function.
The dynamic nature of metabolomic profiles enables real-time monitoring of therapeutic responses and adverse effects. Changes in metabolite concentrations can indicate drug efficacy, toxicity, or off-target effects within hours or days of treatment initiation. This temporal resolution makes metabolomics particularly valuable for therapeutic monitoring and dose optimization in clinical practice.
Integration of metabolomics with other omic data types reveals mechanistic connections between genetic variants, gene expression changes, and functional outcomes. Metabolomic pathway analysis can identify dysregulated biochemical processes that suggest specific therapeutic interventions. Multi-omic data integration precision medicine uses metabolomic data to validate hypotheses generated from genomic, transcriptomic, and proteomic analyses while providing functional context for molecular observations.
Pathology: Spatial and Morphological Context
Digital pathology and spatial omics technologies add critical spatial and morphological dimensions to multi-omic data integration precision medicine approaches. Traditional histopathological analysis provides essential information about tissue architecture, cellular morphology, and disease staging that cannot be captured through molecular analyses alone. Digital pathology platforms enable quantitative analysis of these features while maintaining the spatial context that is lost in homogenized molecular samples.
Spatial transcriptomics and proteomics technologies enable measurement of molecular profiles while preserving spatial organization within tissues. These approaches reveal how molecular heterogeneity relates to anatomical structures and cellular interactions. The integration of spatial molecular data with traditional pathology enhances understanding of disease mechanisms and progression patterns.
Multi-omic data integration precision medicine benefits significantly from pathological context, as morphological features often correlate with molecular subtypes and clinical outcomes. The combination of quantitative pathology with molecular profiling enables more accurate diagnosis, prognosis, and treatment selection than either approach alone. Artificial intelligence applications in pathology can identify morphological patterns that correlate with molecular features, creating integrated diagnostic algorithms.
 Technology Evolution Timeline showing convergence of omic technologies
Technology Evolution Timeline showing convergence of omic technologies
Computational Challenges and Solutions
The successful implementation of multi-omic data integration precision medicine requires sophisticated computational approaches that can handle diverse data types, scales, and quality characteristics. Data harmonization represents a fundamental challenge, as different omic technologies produce data with varying scales, distributions, and error structures. Standardization protocols and normalization methods must account for technical variations while preserving biological signals.
Machine learning and artificial intelligence approaches are increasingly important for extracting meaningful patterns from complex multi-omic datasets. Deep learning architectures can model non-linear relationships between molecular features and clinical outcomes while handling missing data and technical artifacts. Network-based approaches reveal functional relationships between molecular components across different omic layers.
Cloud computing and federated analysis platforms enable large-scale multi-omic studies while addressing data privacy and security concerns. These technologies allow researchers to analyze distributed datasets without centralizing sensitive information. Multi-omic data integration precision medicine benefits from collaborative platforms that enable data sharing while maintaining appropriate access controls and privacy protections.
Clinical Applications and Translation
The translation of multi-omic data integration precision medicine from research settings to clinical practice requires robust validation, standardization, and regulatory approval processes. Clinical laboratory standards for multi-omic testing must ensure analytical validity, clinical utility, and cost-effectiveness. Quality control measures must address the unique challenges of integrating diverse data types with different error profiles and technical requirements.
Oncology represents the most advanced clinical application area for multi-omic approaches, with tumor profiling increasingly incorporating genomic, transcriptomic, and proteomic analyses. Comprehensive cancer panels that analyze hundreds of genes simultaneously are becoming standard of care for many tumor types. The integration of circulating tumor DNA analysis, imaging biomarkers, and clinical data creates comprehensive monitoring platforms for treatment response and disease progression.
Rare disease diagnosis benefits significantly from multi-omic approaches, as these conditions often involve complex molecular mechanisms that are not apparent from single-gene analyses. Whole exome and genome sequencing combined with transcriptomic and metabolomic analyses can identify pathogenic variants and functional consequences that enable definitive diagnosis and targeted treatment.
Future Directions and Opportunities
The future of multi-omic data integration precision medicine will be shaped by continued technological advancement, regulatory evolution, and clinical validation. Single-cell multi-omics technologies that simultaneously measure multiple molecular layers within individual cells promise unprecedented resolution of cellular heterogeneity and developmental processes. These approaches will enhance understanding of disease mechanisms and treatment responses at the cellular level.
Real-time monitoring technologies that combine wearable sensors, liquid biopsies, and point-of-care testing will enable continuous assessment of molecular profiles and clinical status. The integration of these dynamic measurements with static genomic information will create personalized health monitoring systems that can predict and prevent adverse events before they occur.
Global collaboration and data sharing initiatives will accelerate the development and validation of multi-omic approaches while ensuring that precision medicine benefits reach diverse populations. International consortiums and federated analysis platforms will enable large-scale studies that would be impossible for individual institutions while maintaining appropriate privacy and security protections.
Multi-omic data integration precision medicine represents the convergence of multiple technological and scientific advances that promise to transform healthcare delivery. Success in this field requires interdisciplinary collaboration between clinicians, researchers, bioinformaticians, and regulatory specialists who can navigate the complex challenges of translating multi-dimensional molecular insights into improved patient outcomes. The organizations and healthcare systems that successfully implement these approaches will be positioned to deliver truly personalized medicine that optimizes outcomes while minimizing costs and adverse effects.